Sex Check Module¶
The usage of the standalone module is shown below:
$ pyGenClean_sex_check --help
usage: pyGenClean_sex_check [-h] [-v] --bfile FILE [--femaleF FLOAT]
[--maleF FLOAT] [--nbChr23 INT] [--gender-plot]
[--sex-chr-intensities FILE]
[--gender-plot-format FORMAT] [--lrr-baf]
[--lrr-baf-raw-dir DIR] [--lrr-baf-format FORMAT]
[--out FILE]
Check sample's gender using Plink.
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exit
Input File:
--bfile FILE The input file prefix (will find the Plink binary
files by appending the prefix to the .bed, .bim, and
.fam files, respectively.
Options:
--femaleF FLOAT The female F threshold. [default: < 0.300000]
--maleF FLOAT The male F threshold. [default: > 0.700000]
--nbChr23 INT The minimum number of markers on chromosome 23 before
computing Plink's sex check [default: 50]
Gender Plot:
--gender-plot Create the gender plot (summarized chr Y intensities
in function of summarized chr X intensities) for
problematic samples.
--sex-chr-intensities FILE
A file containing alleles intensities for each of the
markers located on the X and Y chromosome for the
gender plot.
--gender-plot-format FORMAT
The output file format for the gender plot (png, ps,
pdf, or X11 formats are available). [default: png]
LRR and BAF Plot:
--lrr-baf Create the LRR and BAF plot for problematic samples
--lrr-baf-raw-dir DIR
Directory or list of directories containing
information about every samples (BAF and LRR).
--lrr-baf-format FORMAT
The output file format for the LRR and BAF plot (png,
ps, pdf, or X11 formats are available). [default: png]
Output File:
--out FILE The prefix of the output files (which will be a Plink
binary file. [default: sexcheck]
Input Files¶
This module uses PLINK’s binary file format (bed, bim and fam files)
for the source data set (the data of interest).
If the option of generating the gender plot is used, a file containing intensities information about each markers on the sexual chromosomes is required. This file (which could be gzipped) should contain at least the following columns:
Sample ID: the unique sample id for each individual.SNP Name: the unique name of each markers.Chr: the name of the chromosome on which each marker is located.X: the intensities of the first allele of each marker.Y: the intensities of the second allele of each marker.
If the options of generating the BAF and LRR values is used, the name of a directory containing intensities file for each sample is required. The name of each file should be the unique sample id. This file (which could be gzipped) should contain at least the following columns:
Chr: the name of the chromosome on which each marker is located.Position: the position of the marker on the chromosome.B Allele Freq: the BAF value of each marker.Log R Ratio: the LRR value of each marker.
If the two plotting module is used alone, one more file is required per module:
a list of samples with gender problem and explanation for
pyGenClean.SexCheck.gender_plot, and only the list of samples with
gender problem for pyGenClean.SexCheck.baf_lrr_plot (both files are
provided by the pyGenClean.SexCheck.sex_check module).
Procedure¶
Here are the steps performed by the module:
- Checks if there are enough markers on the chromosome 23. If not, the module stops here.
- Runs the sex check analysis using Plink.
- If there are no sex problems, the module quits.
- Creates the recoded file for the chromosome 23.
- Computes the heterozygosity percentage on the chromosome 23.
- If there are enough markers on chromosome 24 (at least 1), creates the recoded file for this chromosome.
- Computes the number of no call on the chromosome 24.
- If required, plots the gender plot.
- If there are
summarized_intensitiesprovided, reads the files and skips to step vi. - Reads the
bimfile to get markers on the sexual chromosomes. - Reads the
famfile to get individual’s gender. - Reads the file containing samples with sex problems.
- Reads the intensities and summarizes them.
- Plots the summarized intensities.
- If there are
- If required, plots the BAF and LRR plots.
- Reads the problematic samples.
- Finds and checks the raw files for each of the problematic samples.
- Plots the BAF and LRR plots.
Output Files¶
The output files of each of the steps described above are as follow (note that
the output prefix shown is the one by default [i.e sexcheck]):
- No output files created.
- One set of PLINK’s result files:
sexcheck: the result of the sex check procedure from Plink.
- Two files are created if there are sex problems:
sexcheck.list_problem_sex: a summary of samples with sex problem.sexcheck.list_problem_sex_ids: the list of sample ids with sex problem.
- One set of Plink’s files:
sexcheck.chr23_recodeA: the recoded file for the chromosome 23.
- One custom output file:
sexcheck.chr23_recodeA.raw.hetero: the heterozygosity percentage on the chromosome 23. The file includes the following columns:PED(the pedigree ID),ID(the individual ID),SEX(the gender) andHETERO(the heterozygosity).
- One set of Plink’s files:
sexcheck.chr24_recodeA: the recoded file for the chromosome 24.
- One custom output file:
sexcheck.chr24_recodeA.raw.noCall: the number of no call on the chromosome 24. The file includes the following columns:PED(the pedigree ID),ID(the individual ID),SEX(the gender),nbGeno(the number of genotypes on the chromosome 24) andnbNoCall(the number of genotypes that were not called on chromosome 24).
- Multiple files and one plot. The files are created so that the plot can be
generated again with different parameters (since the summarized intensities
for each sample are really long to compute).
sexcheck.png: the gender plot (see Figure Gender plot).sexcheck.ok_females.txt: the list of females without sex problem, including their summarized intensities on chromosome 23 and 24.sexcheck.ok_males.txt: the list of males without sex problem, including their summarized intensities on chromosome 23 and 24.sexcheck.ok_unknowns.txt: the list of unknown gender without sex problem, including their summarized intensities on chromosome 23 and 24.sexcheck.problematic_females.txt: the list of females with sex problem, including their summarized intensities on chromosome 23 and 24.sexcheck.problematic_males.txt: the list of males with sex problem, including their summarized intensities on chromosome 23 and 24.sexcheck.problematic_unknowns.txt: the list of unknown gender with sex problem, including their summarized intensities on chromosome 23 and 24. When this file is created by thepyGenClean.SexCheck.sex_checkmodule, it is empty.
- A directory containing one file per individual with gender problem (see Figure BAF and LRR plot).
The Plots¶
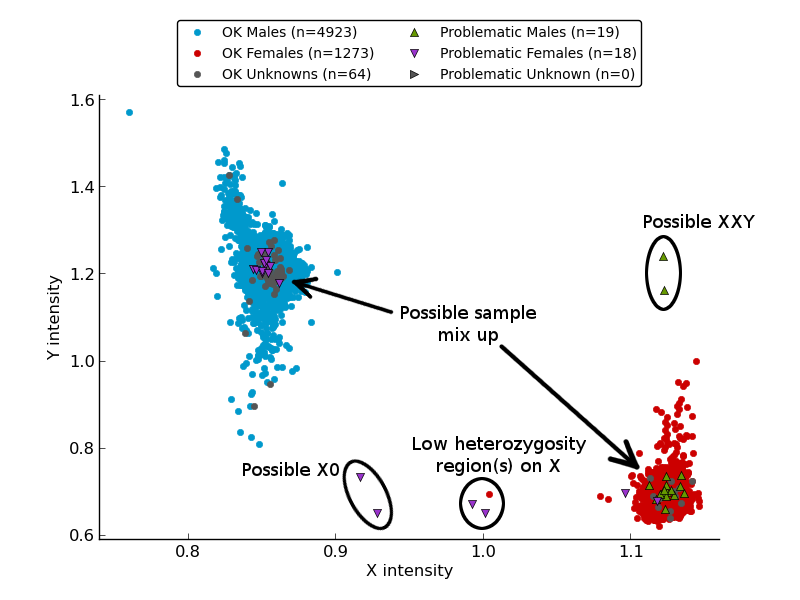
The plot generated by the pyGenClean.SexCheck.gender_plot module (the
Gender plot figure) represents the summarized intensities of each
sample of the data set. The color code represent the gender of each sample. Blue
and red dots represent the males and females without gender problem,
respectively. The green and purple triangles represent the males and females
with gender problem. The gray dots and triangles represent the samples with
unknown gender, with and without gender problem, respectively. This plot makes
possible to find samples with sexual chromosomes abnormalities, such as males
which are XXY or females with only one copy of the X chromosome (X0). Males that
appear to be females and vice versa might in fact be sample mix up and would
require further analysis.
The Gender plot figure can also be manually created after the data clean up pipeline, using its results and this following standalone script:
$ pyGenClean_gender_plot --help
usage: pyGenClean_gender_plot [-h] [-v] --bfile FILE [--intensities FILE]
[--summarized-intensities FILE] --sex-problems
FILE [--format FORMAT] [--xlabel STRING]
[--ylabel STRING] [--out FILE]
Plots the gender using X and Y chromosomes' intensities
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exit
Input File:
--bfile FILE The plink binary file containing information about
markers and individuals. Must be specified if
'--summarized-intensities' is not.
--intensities FILE A file containing alleles intensities for each of the
markers located on the X and Y chromosome. Must be
specified if '--summarized-intensities' is not.
--summarized-intensities FILE
The prefix of six files (prefix.ok_females.txt,
prefix.ok_males.txt, prefix.ok_unknowns.txt,
problematic_females.txt, problematic_males.txt and
problematic_unknowns.txt) containing summarized chr23
and chr24 intensities. Must be specified if '--bfile'
and '--intensities' are not.
--sex-problems FILE The file containing individuals with sex problems.
This file is not read if the option 'summarized-
intensities' is used.
Options:
--format FORMAT The output file format (png, ps, pdf, or X11 formats
are available). [default: png]
--xlabel STRING The label of the X axis. [default: X intensity]
--ylabel STRING The label of the Y axis. [default: Y intensity]
Output File:
--out FILE The prefix of the output files (which will be a Plink
binary file. [default: sexcheck]
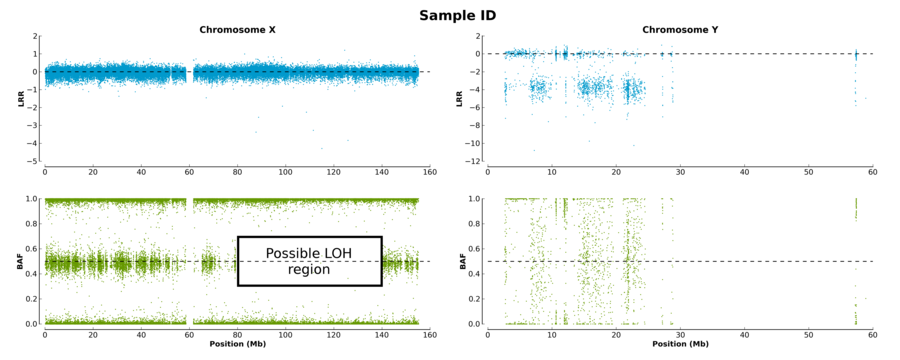
The log R ratio (LRR) for a sample is the log ratio of the normalized R value for the marker divided by the expected normalized R value. Hence, a value of 0 means 2 copies. A drop in the LRR shows a loss of a copy, while an increasing LRR shows a gain of a copy. Expected LRR values on chromosome 23 for female and female are 0 and [-0.5, -1], respectively. The LRR values of each markers on both the X and Y chromosomes are shown in the BAF and LRR plot figure.
The B allele frequency (BAF) for a sample shows the theta value for a marker, corrected for cluster position. For a normal number of copies, markers should have a BAF around 1 (homozygous for the B allele), 0.5 (heterozygous) or 0 (homozygous for A allele). Normal females should have the three lines across the chromosome. Normal males should only have two lines, located near 1 or 0. The BAF values of each markers on both the X and Y chromosomes are shown in the BAF and LRR plot figure.
The BAF and LRR plot figure can also be manually created after the data clean up pipeline, using this following standalone script:
$ pyGenClean_baf_lrr_plot --help
usage: pyGenClean_baf_lrr_plot [-h] [-v] --problematic-samples FILE
[--use-full-ids] [--full-ids-delimiter CHAR]
--raw-dir DIR [--format FORMAT] [--out FILE]
Plots the BAF and LRR of problematic samples.
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exit
Input File:
--problematic-samples FILE
A file containing the list of samples with sex
problems (family and individual ID required, separated
by a single tabulation). Uses only the individual ID
by default, unless --use-full-ids is used.
--use-full-ids Use this options to use full sample IDs (famID and
indID). Otherwise, only the indID will be use.
--full-ids-delimiter CHAR
The delimiter between famID and indID for the raw file
names. [default: _]
--raw-dir DIR Directory containing information about every samples
(BAF and LRR).
Options:
--format FORMAT The output file format (png, ps, pdf, or X11 formats
are available). [default: png]
Output File:
--out FILE The prefix of the output files. [default: sexcheck]
The Algorithm¶
For more information about the actual algorithms and source codes, refer to the following pages.