Windows Installation¶
The following steps will help you install pyGenClean on a Windows machine. It has been tested on both Windows XP and Windows 7.
Requirements¶
The following softwares and packages are required for pyGenClean:
- Python 2.7
- PLINK (1.07)
numpy(version 1.6.2 or latest)matplotlib(version 1.2.0 or latest)scipy(version 0.11.0 or latest)scikit-learn(version 0.12.1 or latest)Jinja2(version 2.7.3 or latest)
Note
All the requirements will be installed along with the main
pyGenClean module.
Warning
The Plink software needs to be in the PATH (or in the current working directory). In other words, you should be able to type plink at the command line.
Installation¶
The easiest way to install Python on windows is by using Miniconda.
Miniconda¶
Download and install miniconda (located at http://conda.pydata.org/miniconda.html).
To create a new virtual environment, perform the following command:
$ conda create -n Python-2.7_virtualenv python=2
Activating the conda environment¶
To activate the Python virtual environment (miniconda), perform the following command:
$ activate Python-2.7_virtualenv
Installing pyGenClean¶
To install pyGenClean, only perform the following command:
$ conda install pyGenClean -c http://statgen.org/wp-content/uploads/Softwares/pyGenClean
Updating pyGenClean¶
To update pyGenClean, perform the following command:
$ conda update pyGenClean -c http://statgen.org/wp-content/uploads/Softwares/pyGenClean
Testing the Algorithm¶
To test the algorithm, download the test data from
http://statgen.org/downloads/pygenclean/ and the HapMap reference
populations (build 37). Create a directory on your Desktop named
pyGenClean_test, and extract the two archive into it. You should have the
following directory structure:
Desktop\
pyGenClean_test_data\
1000G_EUR-MXL_Human610-Quad-v1_H.bed
1000G_EUR-MXL_Human610-Quad-v1_H.bim
1000G_EUR-MXL_Human610-Quad-v1_H.fam
check_ethnicity_HapMap_ref_pops_b37\
hapmap_CEU_r23a_filtered_b37.bed
hapmap_CEU_r23a_filtered_b37.bim
hapmap_CEU_r23a_filtered_b37.fam
hapmap_YRI_r23a_filtered_b37.bed
hapmap_YRI_r23a_filtered_b37.bim
hapmap_YRI_r23a_filtered_b37.fam
hapmap_JPT_CHB_r23a_filtered_b37.bed
hapmap_JPT_CHB_r23a_filtered_b37.bim
hapmap_JPT_CHB_r23a_filtered_b37.fam
Open the command prompt and navigate to the newly created directory, and
created an new text file using notepad:
> cd Desktop\pyGenClean_test
> notepad conf.ini
Insert the following code in the file:
1 2 3 4 5 6 7 8 9 10 | [1]
script = check_ethnicity
ceu-bfile = check_ethnicity_HapMap_ref_pops_b37/hapmap_CEU_r23a_filtered_b37
yri-bfile = check_ethnicity_HapMap_ref_pops_b37/hapmap_YRI_r23a_filtered_b37
jpt-chb-bfile = check_ethnicity_HapMap_ref_pops_b37/hapmap_JPT_CHB_r23a_filtered_b37
nb-components = 2
multiplier = 1
[2]
script = sex_check
|
Finally, run the following command:
> run_pyGenClean ^
--conf conf.ini ^
--bfile pyGenClean_test_data\1000G_EUR-MXL_Human610-Quad-v1_H
Results¶
Valuable information will be shown on the command prompt. Once the program has
finished, the results are in the new directory data_clean_up.date_time
where date is the current date, and time is the time when the program
started.
Here are the new directory structure, with only the files you might be interested in:
data_clean_up.data_time\pyGenClean.logautomatic_report.texexcluded_markers.txtexcluded_samples.txt1_check_ethnicity\ethnicity.before.pngethnicity.outliers.pngethnicity.outliersethnicity.population_file_outliers
2_sex_check\sexcheck.list_problem_sex
The file pyGenClean.log contain the information that was displayed in the
console. The file automatic_report.txt contain the automatic report
generated by pyGenClean. The files excluded_markers.txt and
excluded_samples.txt contains the list of markers and samples,
respectively, that were excluded from the dataset (with the reason).
1_check_ethnicity/¶
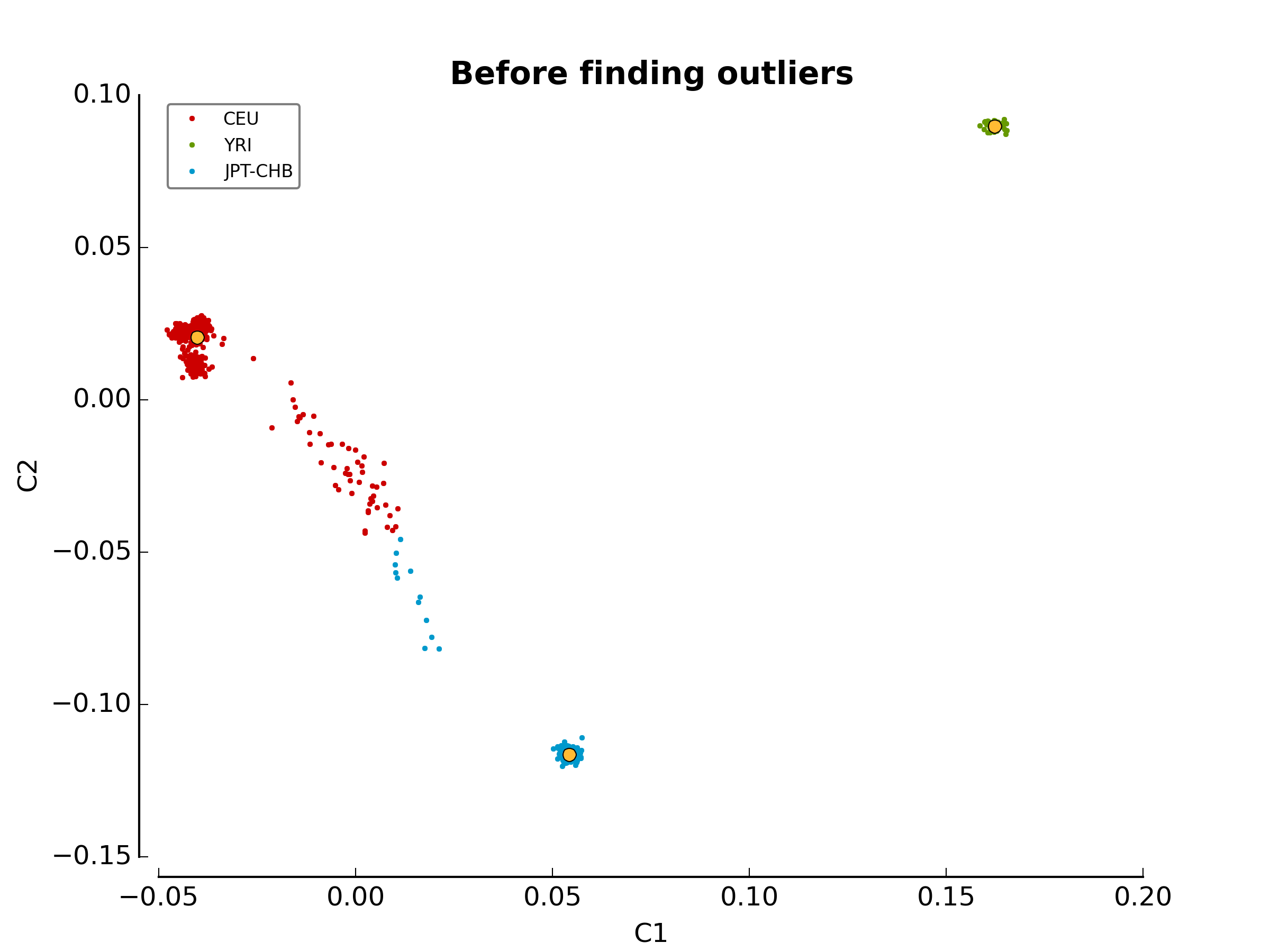
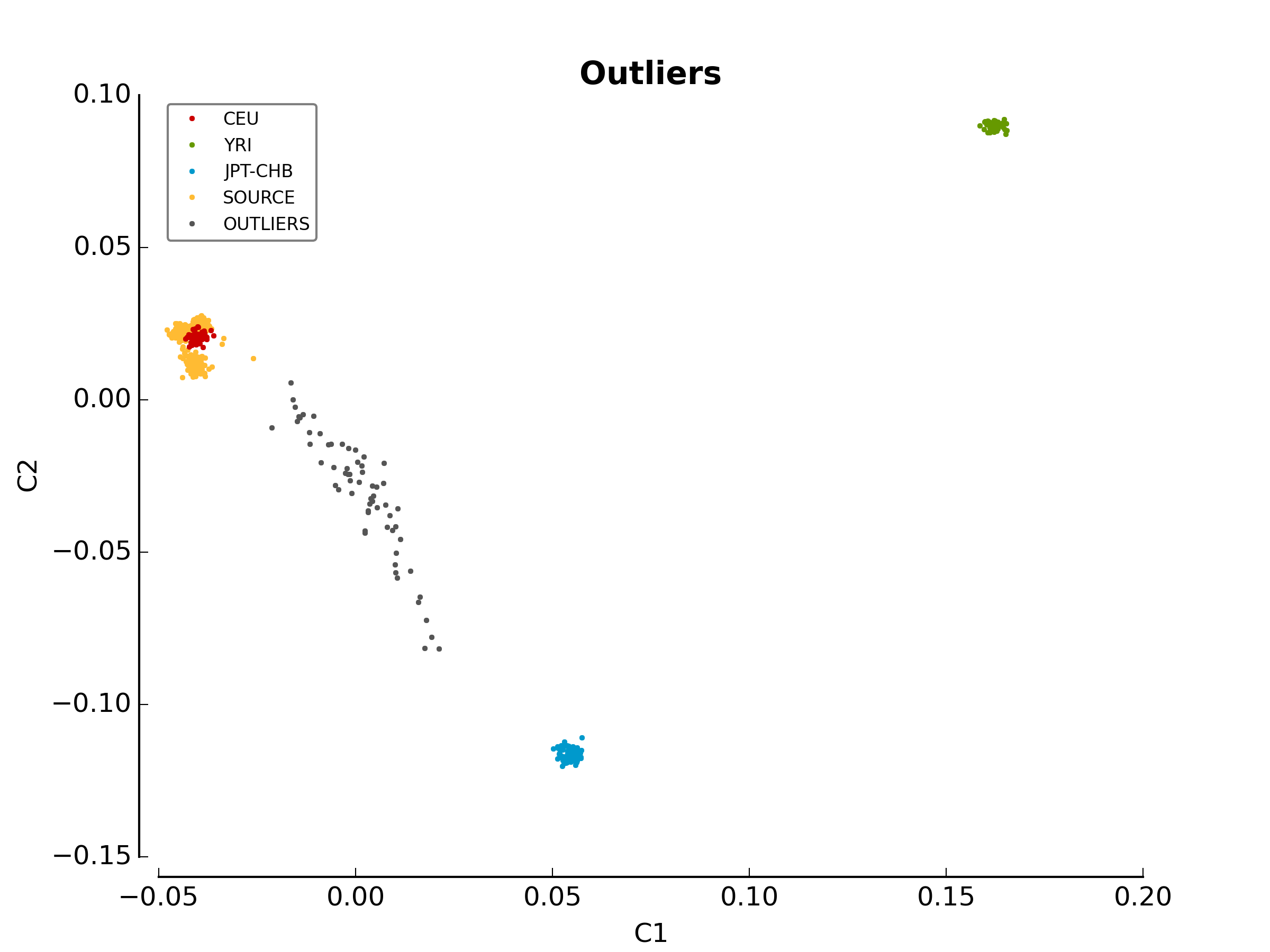
The first image in the first directory
(ethnicity.before.png) shows the MDS values for each sample
before outlier detection. The second image
(ethnicity.outliers.png) shows the outliers that should be
removed for further analysis. Finally, the file ethnicity.outliers include
a list of samples that should be removed for further analysis. The total
number of outliers for this test should be exactly 63. For more information
about the results of this module, refer to Section
Ethnicity Module.
2_sex_check/¶
In the second directory, there should be a file containing the list of samples with gender problem. There should be exactly 4 samples with gender problem. For more information about this module, refer to Section Sex Check Module.
If you want to compare your results with the expected ones, just download the
files in the archive pyGenClean_expected_results.tar.bz2, available through
http://statgen.org/downloads/pygenclean/. They were generated using Fedora
18 (64 bits) in about 20 minutes. You should at least compare the following
files:
1_check_ethnicityethnicity.outliersethnicity.population_file_outliers- All the figures (they might be mirrored).
2_sex_checksexcheck.list_problem_sexsexcheck.list_problem_sex_ids
Automatic report¶
If LaTeX is installed, you can perform the following commands to compile the automatic report into a PDF file.
$ pdflatex automatic_report.tex
$ pdflatex automatic_report.tex
$ pdflatex automatic_report.tex
The following PDF report will be generated.